SIO and DASF for N Estimation¶
This tutorial shows you how to use the Spectral Index Optimizer (SIO) and the DASF retrieval tools of the EnMAP-Box to estimate leaf nitrogen concentration for a hyperspectral satellite image of a forested area.
Aims:
- Get familiar with the EnMAP-Box by applying a simple but meaningful workflow.
- Use the SIO to calculate a regression between spectral data and a variable of interest (here: foliar N concentration)
- Understand basics of the Directional Area Scattering Factor (DASF)
- Calculate the DASF using the tool in the EnMAP-Box
- Calculate the Canopy Scattering factor (CSC) using the ImageMath application in the EnMAP-Box
Introduction¶
Spectral indices (or vegetation indices) have been used for regression and classification problems in remote sensing for decades. With multispectral satellite data like Landsat TM a manageable amount of indices could be calculated from the few available bands.
Hyperspectral data like EnMAP images offer hundreds of bands and thus tens or hundreds of thousands of possible band combinations to calculate simple two-band spectral indices. To find the optimal index for your problem, the Spectral Index Optimizer (SIO) can test all possible two-band combinations (as Normalized Difference Index, Difference Index, or Ratio Index), calculate regressions between each index and your variable of interest and calculate a performance measure (RMSE, MAE, or R^2) for every case. A two-dimensional map of the performance measure is output so you can see spectral regions with high or low correlation to your variable.
In the first part of this tutorial we will use SIO (see also SIO help) on hyperspectral reflectance data to estimate foliar N concentration. Instead of the hyperspectral reflectance, you can use other multi-band images to estimate your variable of interest using the SIO. The second part of the tutorial introduces the Canopy Scattering factor (CSC) as an alternative.
We use an EnMAP data set simulated from an airborne hyperspectral data set recorded with a HySpex VNIR1600 and SWIR320m-e camera. The data set was recorded over the Gerolstein area in Germany on 2016-09-08. The full high resolution airborne data set will soon be available at enmap.org/flights.html.
SIO for N Estimation¶
Preparation¶
- Download the dataset “Tutorial.zip” from http://b8m.de/EnMAP/Tutorial.zip
- Unpack the data, remember the target folder. There should be two images in Envi format consisting of two files each (.bsq and .hdr, see image below).
- Start the EnMAP-Box (please check the EnMAP-Box tutorials first if you don’t know how). You can close all pre-loaded data sources since they aren’t needed for this tutorial.
- Add data sources Gerolstein_2016-09-08_EnMAP.bsq and Gerolstein_2016-09-08_EnMAP_N-Training.dat.
Use SIO tool¶
Open the SIO tool:
- If you don’t see the Processing Toolbox at the right part of the EnMAP-Box, activate it through
View->Panels->Processing Toolbox - In the Processing Toolbox navigate to
EnMAP-Box->Regression->Spectral Index Optimizer. - Double click to start.
- If you don’t see the Processing Toolbox at the right part of the EnMAP-Box, activate it through
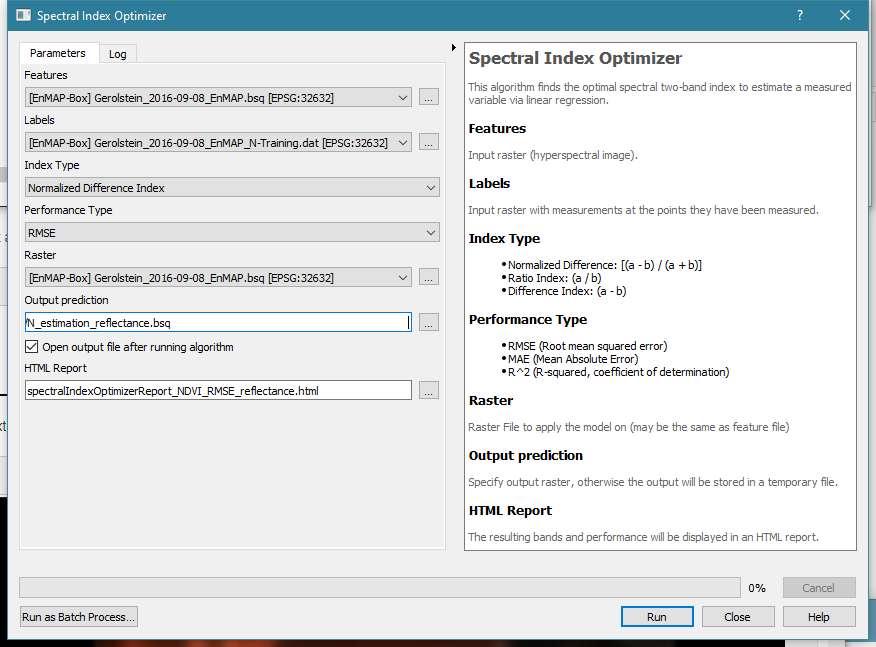
Use the SIO tool to create a map of N estimates:
- In the field
Features, select Gerolstein_2016-09-08_EnMAP.bsq - In the field
Labels, select Gerolstein_2016-09-08_EnMAP_N-Training.dat - You can select any Index Type and Performance Type you like.
- To save the N estimation in a file, click
...at the fieldOutput predictionand enter a path and filename. - Enter a filename for the HTML report or use the default filename.
- Click
Runto run the SIO tool. - It should take only a few seconds, then the HTML report (see below) will open in your browser and the resulting N map will be available in the EnMAP-Box.
- In the field
You can inspect the map in the EnMAP-Box viewer.
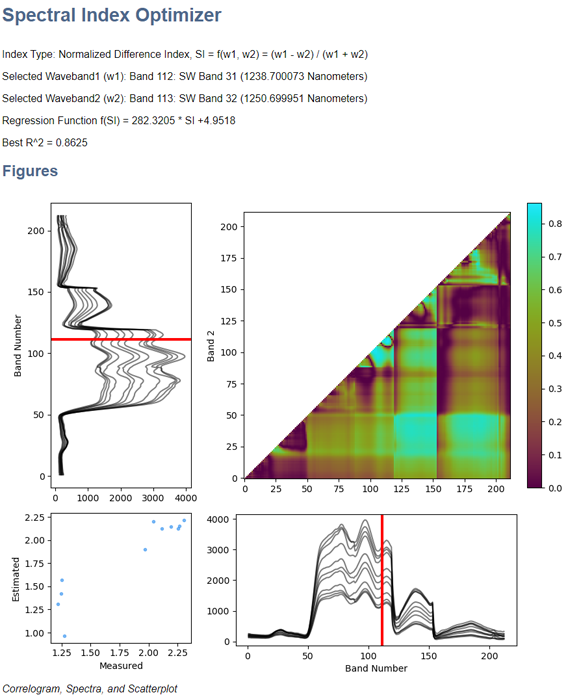
The SIO HTML report looks like this:
Calculate DASF and CSC¶
Broadleaf trees usually have higher leaf N concentrations and appear brighter than needleleaf trees. The brightness can be used to estimate N if both broadleaf and needleleaf trees are in your sample, but the correlation will be spurious (see DASF help text and Knyazikhin et al., 2013).
The Directional Area Scattering Factor (DASF) was invented to compensate for the brightness differences to estimate N from spectra without using the spurious correlation. So in the second part of this tutorial we will calculate the DASF of our image, use it to compensate brightness differences between broadleaf and needleleaf trees and estimate N with the resulting Canopy Scattering Coefficient (CSC) image.
Calculate DASF¶
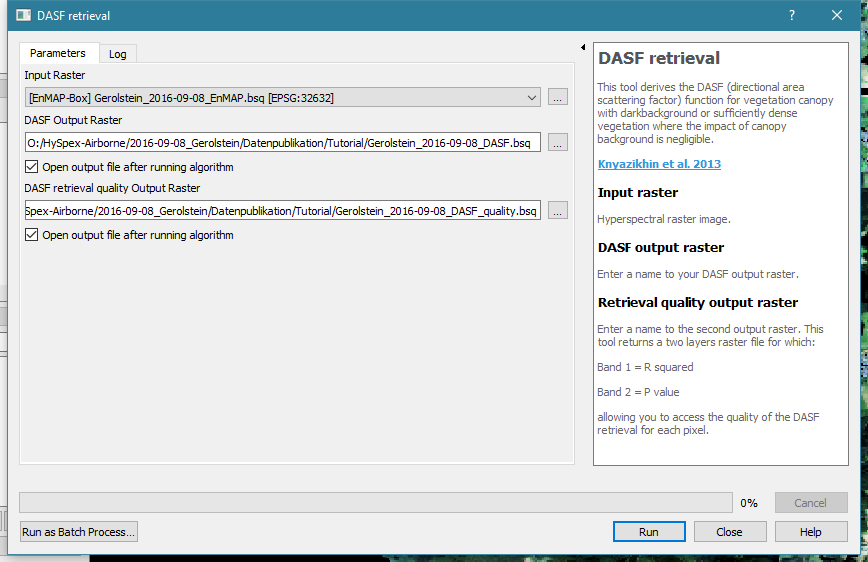
Open the DASF retrieval tool:
- In the Processing Toolbox navigate to
EnMAP-Box->Forest->DASF retrieval - Double click to start.
- In the Processing Toolbox navigate to
Select the hyperspectral reflectance image Gerolstein_2016-09-08_EnMAP.bsq as Input raster.
Specify output files for the DASF output raster and the quality output raster (optional).
Run the DASF retrieval tool. For the tutorial file it should only take about a minute.
The DASF is a single band image that is highly correlated to the bidirectional reflectance in the near infrared region of close forest pixels. Divide the hyperspectral reflectance image by the DASF to get the CSC. In the next step we will use the CSC to estimate N and compare the result to the estimation based on reflectance.
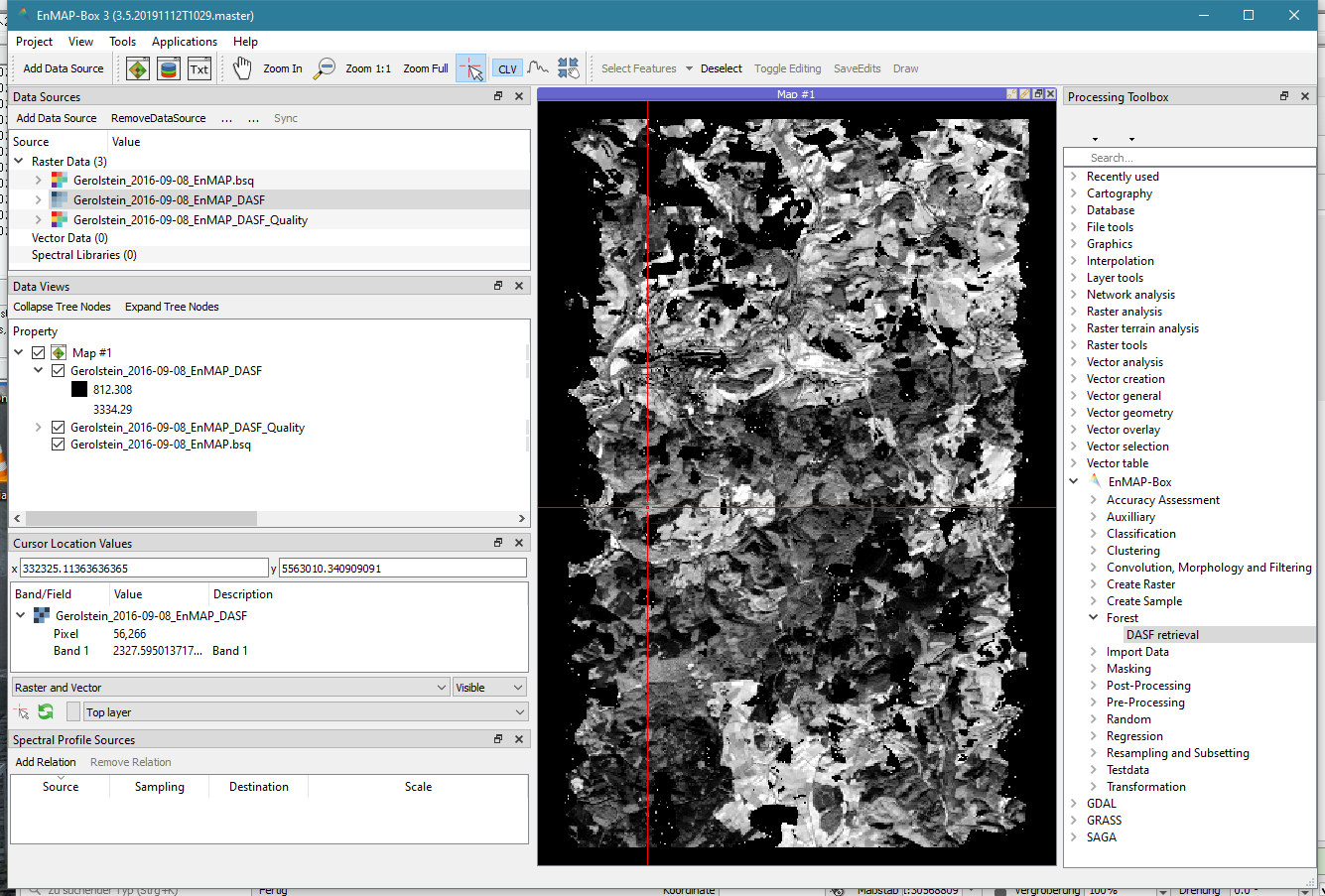
Calculate CSC¶
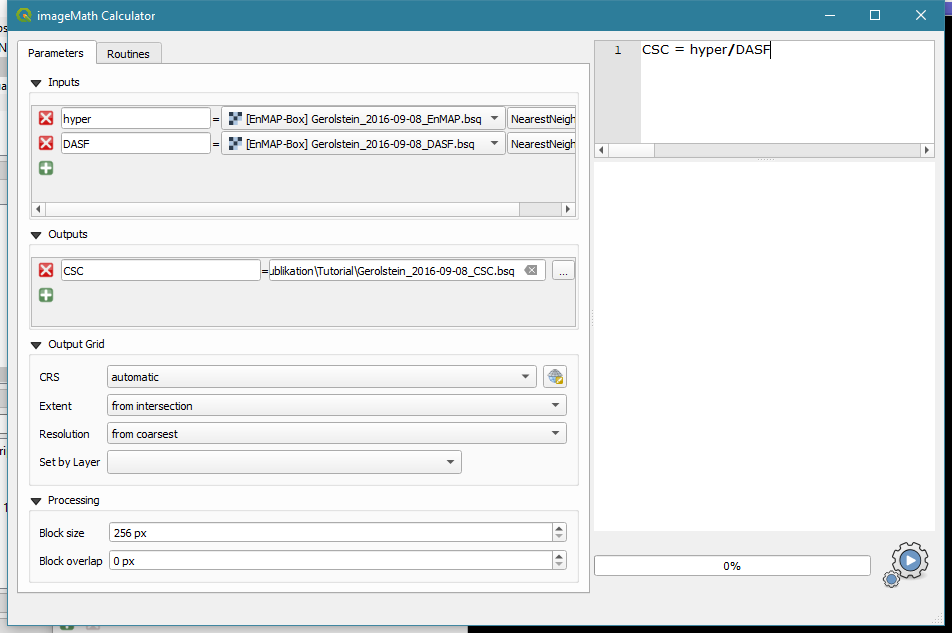
We will use the ImageMath application to divide the reflectance image by the DASF.
- Open the ImageMath application: Navigate to
Applications->ImageMath - In the ImageMath Calculator we need to define input and output variables and an equation to create the output from the input.
- In the grey field under “Inputs”, select the hyperspectral image and call it “hyper” in the white field at the left hand side.
- Click on the green
+to create a second input - Select the DASF and call it DASF
- Under “Outputs”, select a filename for the CSC and call it CSC
- Enter the equation “CSC = hyper/DASF” in the top right field. It uses the variable names we just defined in the Input and Output fields.
- Start the calculation by clicking the gearwheel at the bottom right.
Estimate N using CSC¶
The Nitrogen concentration can be estimated from CSC analogous to the estimation from reflectance:
- In the Processing Toolbox navigate to
EnMAP-Box->Regression->Spectral Index Optimizer, double click to start. - In the field
Features, select the CSC image. - In the field
Labels, select Gerolstein_2016-09-08_EnMAP_N-Training.dat again. - You can select any Index Type and Performance Type you like.
- To save the N estimation in a file, click
...at the fieldOutput predictionand enter a path and filename. - Enter a filename for the HTML report or use the default filename.
- click
Runto run the SIO tool.
Discussion¶
Please use the results with caution.
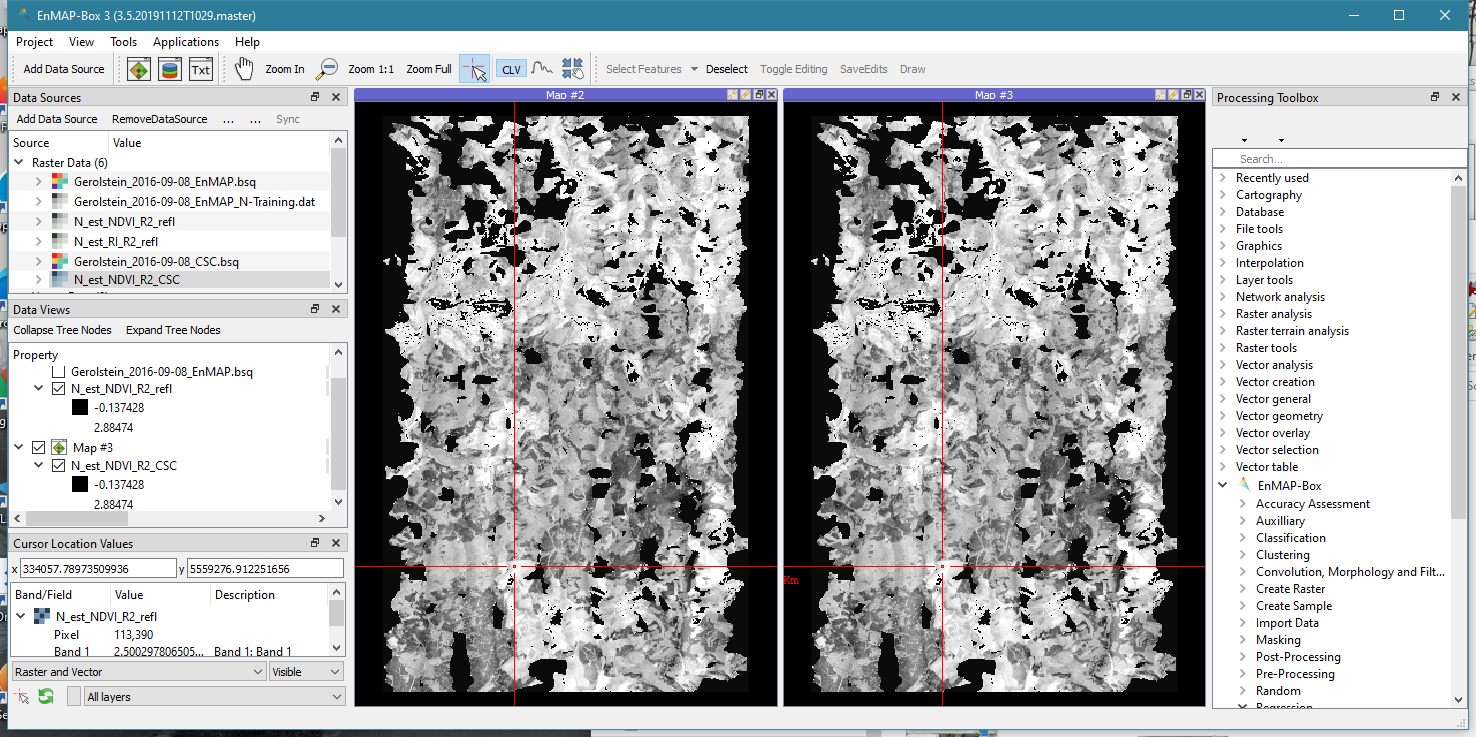
Using the tutorial data, the correlograms will be nearly identical, no matter whether reflectance or CSC data is used. Consequently, the N maps will also be very similar (see image below). This may be different when different data sets are used.
The data set used for this tutorial is based on a field campaign with professional tree climbers who collected leaf samples. These samples were analysed in a laboratory. Unfortunately, only a very low number of samples can be used, because only few samples could be collected in the given time and some sampling points are located in gaps or areas with cloud shadows in the hyperspectral image. This limited data set does not allow for a reliable wall-to-wall estimation of N concentration.
In addition, the N estimation only makes sense for forest pixels, but the image contains many additional land use classes. Masking non-forest pixels is not part of the scope of this tutorial for now.
References¶
Knyazikhin, Y., Schull, M.A., Stenberg, P., Mõttus, M., Rautiainen, M., Yang, Y., Marshak, A., Latorre Carmona, P., Kaufmann, R.K., Lewis, P., Disney, M.I., Vanderbilt, V., Davis, A.B., Baret, F., Jacquemoud, S., Lyapustin, A. & Myneni, R.B. (2013). Hyperspectral remote sensing of foliar nitrogen content. Proceedings of the National Academy of Sciences, 110, E185-E192. DOI:10.1073/pnas.1210196109, http://www.pnas.org/content/110/3/E185.abstract